Tomato Genome Sequence
| Determining the minimum tilling path from the tomato BAC library in order to sequence chromosome 7
In collaboration with:
Website : http://gbf.ensat.fr/gbf_projects.html |
|
Abstract
The tomato genome will be sequenced as the cornerstone of an International Solanaceae Genome Initiative, a project that aims to develop the family Solanaceae as a model for systems biology for understanding plant adaptation and diversification.
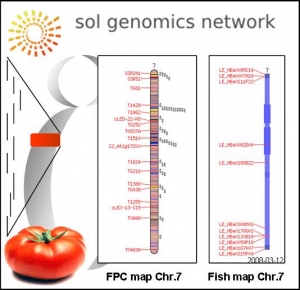
The tomato genome is comprised of approximately 950 Mb of DNA – more than 75% of which is heterochromatin and largely devoid of genes. The majority of genes are found in long contiguous stretches of gene-dense euchromatin located on the distal portions of each chromosome arm. The strategy is to identify and sequence a minimal tiling path of BAC clones through this approximately 220 Mb euchromatin.
The gene-rich euchromatic portion of the tomato genome is being sequenced by an international consortium and the GBF laboratory is leading the French effort for sequencing tomato chromosme 7.
Links: http://www.sgn.cornell.edu/help/about/tomato_sequencing.pl
Results
The first sequencing of the tomato genome has been achieved by the International Tomato Sequencing Consortium and is now available on the SGN web site ( http://solgenomics.net/tomato/).
CNRGV's responsible
- Sonia VAUTRIN
CNRGV's involvement
- Macroarray production/ screening
- 3D DNA pools production
- DNA pools screening
Publication related to the project