PHYTOSOL-2
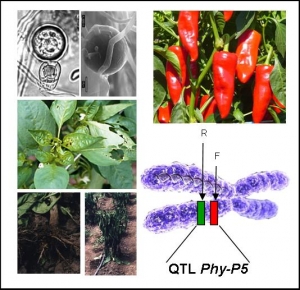
Functional confirmation of candidate genes for a broad-spectrum resistance QTL against Phytophthora in Solanaceae
ANR project- Plant Genomic Network - 2010 Génoplante
Project coordinator :
Dr. Véronique Lefebvre
INRA-UR1052-GAFL
Caractérisation Fonctionnelle des interactions Plantes - Bioagresseurs
BP94 - 84140 Montfavet - France
E.mail: veronique.lefebvre@avignon.inra.fr
Web site:
Abstract :
Phytophthora species are the most devastating pathogens of Dicotyledons. Chemical treatments are banned by the European Commission. Monogenic resistances are often overcome. Polygenic resistances were demonstrated to be more durable, but molecular bases of resistance QTLs are yet unknown.We aim to investigate the molecular nature of a resistance QTL that target several Phytophthora species infecting Solanaceae. Phy-P5 QTL confers a high resistance level to several P. capsici isolates in pepper, and is colinear to resistance QTLs against P. infestans in tomato and potato. The genetic dissection of Phy-P5 locus permitted the identification of two tightly linked QTLs in 0.57 cM . These two loci were then delimited to two single BAC clones that will be sequenced, annotated and anchored to the genetic map, in order to select candidate genes during 2007.The project PHYTOSOL-2 aims to validate candidate genes identified for Phy-P5 QTLs, and to develop a model system to achieve the functional characterization of genes involved in Phytophthora/Solanaceae interactions. The candidate genes will be sequenced in parental lines and different pepper accessions to look for natural variations (EcoTILLING). Analysis of intragenic recombinant plants and sequence comparison of chimeric alleles will help to identify functional domains. Candidate gene validation will be performed by hairy-root transformation on pepper, and by ectopic expression in P. capsici-susceptible tomato. mRNA and protein spatiotemporal expression patterns and the identification of downstream transduction pathways will be performed in pepper and/or tomato depending of the outcome of WP1-WP2. The tomato TILLING platform will be exploited as a reverse genetic tool for functional analysis of the identified genes. At last, the data produced during this project will be used to improve the breeding for resistance to Phytophthora in Solanaceae species.
Technical approaches
Responsible : Sonia Vautrin
- Construction of Capsicum annum HD208 2D pools
- DNA pools screening by RT-PCR
- BACs Ends sequencing
- QTL phy-P5 physical mapping of the BACs clones
Partners
Unité de Génétique et Amélioration des Fruits et Légumes
INRA-GAFL, UR1052
84143 Montfavet CedexManaging Director : Mathilde Causse
Scientist in charge of the project : Véronique Lefebvre
Unité de Recherche en Génomique Végétale
URGV, UMR1165
INRA-CNRS-Univ
91057 Evry CedexManaging Director : Michel Caboche
Scientist in charge of the project : Abdelhafifid Bendahmane
Centre National de Ressources Génomiques Végétales
INRA-CNRGV, UR1258
31326 Castanet Tolosan CedexManaging Director / Scientist in charge of the project : Hélène Bergès

