Pepper eiF4E family
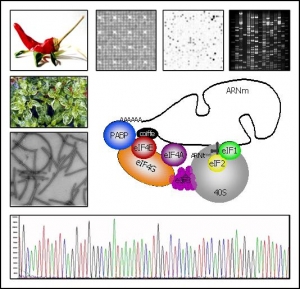
Screening the pepper BAC library in order to identify all the members of the eIF4E multigenic family
In collaboration with
-
Dr. Carole CARANTA
Unité de recherche Génétique et Amélioration des Fruits et Légumes
INRA-UGAFL
BP94 - 84143 Montfavet cedex
France
Website
Abstract
Capsicum resistance to Pepper veinal mottle virus (PVMV) results from complementation between the pvr2 and pvr6 resistance genes: recessive alleles at these two loci are necessary for resistance, whereas any dominant allele confers susceptibility. In line with previous results showing that pvr2 resistance alleles encode mutated versions of the eukaryotic translation initiation factor 4E ( eIF4E), the involvement of other members of the eIF4E multigenic family in PVMV resistance has been investigated. It was demonstrated that pvr6 corresponds to an eIF(iso)4E gene, predicted to encode the second cap-binding isoform identified in plants. Comparative genetic mapping in pepper and tomato indicated that eIF(iso)4E maps in the same genomic region as pvr6.
A Bacterial Artificial Chromosome (BAC) library that consisted of 239,232 clones with an average insert size of 123 kilobases (kb) was constructed from a Capsicum annuum line with the pvr2(+) allele for susceptibility to potato virus Y (PVY) and tobacco etch virus (TEV). Based on a polymerase chain reaction (PCR) screen with single-copy markers, three to seven positive BAC clones per markers were identified, indicating that the BAC library is suitable for pepper genome analysis.
As eIF4E belongs to a small multigenic family in plants, we decided to screen the pepper BAC library in order to identify all the members of this multigenic family.
CNRGV involvement
Responsible : Elisa PRAT
- Macroarray screening
- Fingerprint
- Southern
- eif4E genes sequencing
