
Elisa Prat
Arrival date : 2005
Departure date : 31/10/2024
Status : Laboratory Technician
Roles :
Last completed degree : Graduate of a biochemistry advanced degree and a Bachelor professional of plants biotechnology
- Vincent Castric
- Isabelle Loisy - Thierry Gaude
- Jacinthe Azevedo - Thierry Lagrange
-
Italian National Agency for New technologies, Energy and Environment (ENEA).
ENEA-Casaccia RC, BAS BIOTEC-GEN
Via Anguillarese, 301
00123 Rome
Italy -
Dr. Carole CARANTA
Unité de recherche Génétique et Amélioration des Fruits et Légumes
INRA-UGAFL
BP94 - 84143 Montfavet cedex
France
Website
NOVEL: Emergence of novel phenotypes in co-evolving biological systems: allelic diversification and dominance at the Self-incompatibility locus in Arabidopsis.
Something went wrong when connecting to PubMed API.
Click here to access to the publication on PubMed website

NOVEL: Emergence of novel phenotypes in co-evolving biological systems: allelic diversification and dominance at the Self-incompatibility locus in Arabidopsis.
ERC Consolidator Grant (CoG), LS8, ERC-2014-CoG
Read more
Project coordinator:
Unité Evo-Eco-Paléo (EEP) - UMR 8198
CNRS / Université de Lille - Sciences et Technologies
Batiment SN2, bureau 207
59655 Villeneuve d'Ascq - FRANCE
Project partners:
Laboratory « Reproduction et Developpement des Plantes »
ENS Lyon CNRS - France
Laboratory « Genome et Developpement des Plantes »
UMR 5096 (UPVD/CNRS) - Perpignan - France
Website:
http://eep.univ-lille.fr/fr/perso-vincent-castric
Abstract:
The emerging field of systems biology is revealing the intricate nature of biological organisms, whereby a large fraction of their individual components (genes, proteins, regulatory elements) interact with several others. The co-evolutionary processes that this entails raises the question of how phenotypic novelty may arise in the course of evolution, since all parts of the system have to evolve in a coordinated manner if the phenotype is to remain functional. For most biological systems, however, we are lacking even basic insight into the fine-scale mechanistic constraints and the underlying ecological context. In this project, we will focus on the sporophytic self-incompatibility system in outcrossing Arabidopsis species, a model biological system in which two distinct co-evolutionary processes are becoming well-understood: 1) between the male and female reproductive proteins allowing self-pollen recognition and rejection and 2) between small non-coding RNAs and their target sites that jointly control the dominance/recessivity interactions between self-incompatibility alleles. By studying these two model systems, we will aim to catch the emergence of functional and regulatory novelty in flagrante delicto. We will take a multidisciplinary approach combining theoretical and empirical population genetics, evolutionary genomics and ancestral protein resurrection using transgenic plants. Our goal is threefold: 1) decrypt the molecular alphabet of the interaction between co-evolving nucleotide sequences, 2) predict and evaluate the fitness landscapes upon which the two co-evolutionary processes are taking place and 3) exploit natural variation in closely related species to unveil the kind of co-evolutionary process in natural populations. Our combination of various powerful approaches in a tractable model system should provide insight on diversification, a poorly understood but fundamental evolutionary process that is taking place at all levels of organization.
This project is an extension of the project BRASSIDOM
| CNRGV's involvement: BAC library construction and screening
CNRGV's responsible: William Marande |
|
Publications related to the project:
The unusual S locus of Leavenworthia is composed of two sets of paralogous loci
Evolutionary dynamics and genetic basis of dominance: sporophytic self-incompatibility in the Brassicaceae as a case study.
Something went wrong when connecting to PubMed API.
Click here to access to the publication on PubMed website

Evolutionary dynamics and genetic basis of dominance: sporophytic self-incompatibility in the Brassicaceae as a case study.
Evolutionary dynamics and genetic basis of dominance: sporophytic self-incompatibility in the Brassicaceae as a case study.
Read more
Project coordinator:
Vincent Castric
Unité Evo-Eco-Paléo (EEP) - UMR 8198
CNRS / Université de Lille - Sciences et Technologies
Batiment SN2, bureau 207
59655 Villeneuve d'Ascq - FRANCE
Project partners:
Hélène Touzet at the LIFL - Laboratoire d’Informatique Fondamentale de Lille
CNRS-INRIA- FRANCE
Isabelle Loisy and Thierry Gaude at the laboratory « Reproduction et Developpement des Plantes »
ENS Lyon CNRS - FRANCE
Website:
http://www.agence-nationale-recherche.fr/Project-ANR-11-JSV7-0008
http://eep.univ-lille.fr/fr/perso-vincent-castric
Abstract:
Dominance, the genetic phenomenon whereby one of the two alleles at a diploid locus is masked at the phenotypic level, is one of the earliest observations of classical genetics, yet also one whose genetic basis and evolution remain poorly understood. Dominance modifiers, i.e. genetic elements controlling the dominance of alleles at other genes, have been postulated by Ronald A. Fisher as early as 1928, but they were never documented since then. The question of whether these genetic elements indeed existed and how strongly natural selection could act on them violently opposed him to Sewall Wright in the 30’s.Earlier last year, Tarutani et al. (2010) identified, embedded within the gene cluster controlling self-incompatibility in the Brassicaceae, a trans-acting small RNA acting as dominance modifier of the gene controlling pollen specificity. This discovery provides an elegant mechanistic model for dominance/recessivity interaction in pollen, and hence the first description of dominance modifiers whose existence had been postulated since Fisher,but never documented yet. This opens up the exciting opportunity to address long-standing mechanistic as well as evolutionary issues, such as the type of mutations causing changes in dominance or how intensely natural selection acts on dominance modifiers. In this proposal, we propose to take advantage of this recent discovery and of our previous theoretical and empirical work to shed new light on the evolutionary dynamics and genetic basis of dominance, using an integrative multidisciplinary framework combining (1) phenotypic approaches to describe the dominance network among alleles at the gene controlling pollen self-incompatibility specificity in the plant Arabidopsis halleri; (2) genomic approaches to compare in silico the small RNA repertoire of dominant and recessive haplotypes and validate the proposed mechanism for the control of dominance in a genetic system characterized by strong multiallelism; (3) functional approaches to validate some of the candidate motifs; (4) theoretical approaches to explore how the control of dominance by small RNAs changes our understanding of how dominance can evolve in this genetic system, in particular focusing on the coevolution and asymetry it entails between small RNAs and their targets; (5) molecular evolution approaches to evaluate the level of functional constraint on small RNAs and their target sites.
CNRGV involvement:
BAC library construction and screening
Responsible: William Marande
Publications related to the project:
Screening of wheat genomic BAC libraries with specific primers for the TdDRF1 (Triticum durum Dehydration responsive Factor 1) gene
Something went wrong when connecting to PubMed API.
Click here to access to the publication on PubMed website
Screening of wheat genomic BAC libraries with specific primers for the TdDRF1 (Triticum durum Dehydration responsive Factor 1) gene
Drought is one of the most severe abiotic stresses limiting crop productivity and our understanding, at the molecular level, of crop response to water stress is further increasing.
Read more
In collaboration with:
Website : http://www.enea.it/com/ingl/default.htm
Laboratory Leader: Dr. P. Galeffi
Principal Researcher: Dr. A. Latini
Abstract
The Triticum durum Dehydration Responsive Factor 1 gene, TdDRF1, belonging to the DREB gene family, is expressed in response to dehydration and encodes for abiotic stress responsive transcription factors that impart stress endurance to the plants (Latini et al. 2007). Recently, the expression profile of this gene was finely examined in several wheat varieties in both greenhouse and in field conditions, during a water stress time-course (Latini et al. 2008). All obtained data spotlighted that the targeting of this gene could help the selection of cultivars for improving plant tolerance to drought.
The fine mapping of this gene, determining the exact locus position on chromosomes will be very interesting and useful. Actually, it is already established that homeologous loci of this gene exist in 1A, 1B chromosomes of durum wheat and 1A, 1B and 1D chromosomes of bread wheat; in addiction, there are some evidences about the existence of some paralogous gene copies and also pseudogenes without introns.
The collaboration with the CNRGV is highly fruitful, because it holds the most relevant BAC genomic DNA collections of wheat. In addiction to several collections of bread wheat, at CNRGV, durum wheat (Cenci et al. 2003) and chromosome-specific (such as 1D, 4D, 6D and 1AS or 1AL of hexaploid wheat – Janda et al. 2004) genomic BAC libraries are also available.
Actually, the bread wheat Chinese Spring BAC library (Tae-B-CS) was screened by both PCR on pools and microarray hybridization techniques with primers specific for the TdDRF1 sequence. A durum wheat library is still under screening. Several clones containing the target sequence were identified and are being characterized by restriction analyses, chromosome walking and complete BAC sequencing.
From the positive BAC clones, the upstream and downstream TdDRF1 gene regions will be sequenced, allowing the identification of the promoter and other regulatory sequences (enhancers, alternative splicing signals, and so on). Furthermore, the complete sequence of a selected BAC clone will reveal if some other dehydration-responsive gene is located next to the target locus and this information will be precious for developing applications in molecular assisted breeding programs aimed to the release of drought tolerant wheat cultivars.
- Latini A, Rasi C, Sperandei M, Cantale C, Iannetta M, Dettori M, Ammar K, Galeffi P: Identification of a DREB-related gene in Triticum durum and its expression under water stress conditions. Annals of Applied Biology (2007), 150: 187-195.
- Latini A, Sperandei M, Sharma S, Cantale C, Iannetta M, Dettori M, Ammar K, Galeffi P: Molecular analyses of a dehydration-related gene from the DREB family in durum wheat and triticale. In Biosaline Agriculture and High Salinity Tolerance. Published by Birkhäuser and Verlag. Edited by Abdelly C, Öztürk M, Ashraf M, Grignon C. (2008), 286-295.
- Cenci a, Chantret N, Kong X, Gu Y, Anderson OD, Fahima T, Distelfeld, Dubcovsky J: Construction and characterization of a half million clone BAC library of durum wheat ( Triticum turgidum ssp. durum). Theoretical and Applied Genetics (2003), 107: 931-939.
- Janda J, Bartoš J, Šafář J, Kubaláková M, Valárik M, Čihalíková J, Šimková H, Caboche M, Sourdille P, Bernard M, Chalhoumb B, Doležel J: Construction of a subgenomic BAC library specific for chromosomes 1D, 4D and 6D of Hexaploid wheat. Theoretical and Applied Genetics (2004) 109: 1337-1345.
This collaboration was also supported by a COST-STSM-FA0604 grant, given to A. Latini.
CNRGV involvement
Responsible : Elisa PRAT/ Sonia VAUTRIN
- Macroarray production/ screening
- 2D DNA pools screening
Pepper eiF4E family
Something went wrong when connecting to PubMed API.
Click here to access to the publication on PubMed website
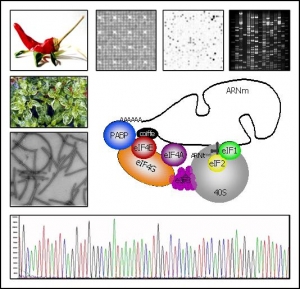
Pepper eiF4E family
Screening the pepper BAC library in order to identify all the members of the eIF4E multigenic family
Read more
Screening the pepper BAC library in order to identify all the members of the eIF4E multigenic family
In collaboration with
Abstract
Capsicum resistance to Pepper veinal mottle virus (PVMV) results from complementation between the pvr2 and pvr6 resistance genes: recessive alleles at these two loci are necessary for resistance, whereas any dominant allele confers susceptibility. In line with previous results showing that pvr2 resistance alleles encode mutated versions of the eukaryotic translation initiation factor 4E ( eIF4E), the involvement of other members of the eIF4E multigenic family in PVMV resistance has been investigated. It was demonstrated that pvr6 corresponds to an eIF(iso)4E gene, predicted to encode the second cap-binding isoform identified in plants. Comparative genetic mapping in pepper and tomato indicated that eIF(iso)4E maps in the same genomic region as pvr6.
A Bacterial Artificial Chromosome (BAC) library that consisted of 239,232 clones with an average insert size of 123 kilobases (kb) was constructed from a Capsicum annuum line with the pvr2(+) allele for susceptibility to potato virus Y (PVY) and tobacco etch virus (TEV). Based on a polymerase chain reaction (PCR) screen with single-copy markers, three to seven positive BAC clones per markers were identified, indicating that the BAC library is suitable for pepper genome analysis.
As eIF4E belongs to a small multigenic family in plants, we decided to screen the pepper BAC library in order to identify all the members of this multigenic family.
CNRGV involvement
Responsible : Elisa PRAT
- Macroarray screening
- Fingerprint
- Southern
- eif4E genes sequencing
The haplotype-resolved chromosome pairs of a heterozygous diploid African cassava cultivar reveal novel pan-genome and allele-specific transcriptome features
Something went wrong when connecting to PubMed API.
Click here to access to the publication on PubMed website
Genome sequencing reveals the origin of the allotetraploid arabidopsis suecica
Something went wrong when connecting to PubMed API.
Click here to access to the publication on PubMed website
Genome sequencing reveals the origin of the allotetraploid arabidopsis suecica
Novikova PY, Tsuchimatsu T, Simon S, Nizhynska V, Voronin V, Burns R, Fedorenko OM, Holm S, Saell T, Prat E, Marande W, Castric V, Nordborg M.
Journal: Molecular Biology and Evolution
DOI: 34: 957-968.
Read more
Novikova PY, Tsuchimatsu T, Simon S, Nizhynska V, Voronin V, Burns R, Fedorenko OM, Holm S, Saell T, Prat E, Marande W, Castric V, Nordborg M.
Journal: Molecular Biology and Evolution
DOI: 34: 957-968.
Abstract
Polyploidy is an example of instantaneous speciation when it involves the formation of a new cytotype that is incompatible with the parental species. Because new polyploid individuals are likely to be rare, establishment of a new species is unlikely unless polyploids are able to reproduce through self-fertilization (selfing), or asexually. Conversely, selfing (or asexuality) makes it possible for polyploid species to originate from a single individual—a bona fide speciation event. The extent to which this happens is not known. Here, we consider the origin of Arabidopsis suecica, a selfing allopolyploid between Arabidopsis thaliana and Arabidopsis arenosa, which has hitherto been considered to be an example of a unique origin. Based on whole-genome re-sequencing of 15 natural A. suecica accessions, we identify ubiquitous shared polymorphism with the parental species, and hence conclusively reject a unique origin in favor of multiple founding individuals. We further estimate that the species originated after the last glacial maximum in Eastern Europe or central Eurasia (rather than Sweden, as the name might suggest). Finally, annotation of the self-incompatibility loci in A. suecica revealed that both loci carry non-functional alleles. The locus inherited from the selfing A. thaliana is fixed for an ancestral non-functional allele, whereas the locus inherited from the outcrossing A. arenosa is fixed for a novel loss-of-function allele. Furthermore, the allele inherited from A. thaliana is predicted to transcriptionally silence the allele inherited from A. arenosa, suggesting that loss of self-incompatibility may have been instantaneous.
Patterns of polymorphism at the self-incompatibility locus in 1,083 Arabidopsis thaliana genomes.
Something went wrong when connecting to PubMed API.
Click here to access to the publication on PubMed website
Patterns of polymorphism at the self-incompatibility locus in 1,083 Arabidopsis thaliana genomes.
Mol Biol Evol. doi: 10.1093/molbev/msx122.
Read more
Authors :
Tsuchimatsu T, Goubet PM, Gallina S, Holl AC, Fobis-Loisy I, Bergès H, Marande W, Prat E, Meng D, Long Q, Platzer A, Nordborg M, Vekemans X, Castric V.
Mol Biol Evol. doi: 10.1093/molbev/msx122.
Abstract :
Although the transition to selfing in the model plant Arabidopsis thaliana involved the loss of the self-incompatibility (SI) system, it clearly did not occur due to the fixation of a single inactivating mutation at the locus determining the specificities of SI (the S-locus). At least three groups of divergent haplotypes (haplogroups), corresponding to ancient functional S-alleles, have been maintained at this locus, and extensive functional studies have shown that all three carry distinct inactivating mutations. However, the historical process of loss of SI is not well understood, in particular its relation with the last glaciation. Here, we took advantage of recently published genomic re-sequencing data in 1,083 Arabidopsis thaliana accessions that we combined with BAC sequencing to obtain polymorphism information for the whole S-locus region at a species-wide scale. The accessions differed by several major rearrangements including large deletions and inter-haplogroup recombinations, forming a set of haplogroups that are widely distributed throughout the native range and largely overlap geographically. 'Relict' A. thaliana accessions that directly derive from glacial refugia are polymorphic at the S-locus, suggesting that the three haplogroups were already present when glacial refugia from the last Ice Age became isolated. Inter-haplogroup recombinant haplotypes were highly frequent, and detailed analysis of recombination breakpoints suggested multiple independent origins. These findings suggest that the complete loss of SI in A. thaliana involved independent self-compatible mutants that arose prior to the last Ice Age, and experienced further rearrangements during post-glacial colonization.
Long Read Sequencing Technology to Solve Complex Genomic Regions Assembly in Plants.
Something went wrong when connecting to PubMed API.
Click here to access to the publication on PubMed website
Long Read Sequencing Technology to Solve Complex Genomic Regions Assembly in Plants.
Journal of Next Generation Sequencing & Applications.
Read more
Authors :
Arnaud Bellec, Audrey Courtial, Stephane Cauet, Nathalie Rodde, Sonia Vautrin, Genseric Beydon, Nadege Arnal, Nadine Gautier, Joelle Fourment, Elisa Prat, William Marande, Yves Barriere and Helene Berges.
Journal of Next Generation Sequencing & Applications.
Abstract :
Background:
Numerous completed or on-going whole genome sequencing projects have highlighted the fact that obtaining a high quality genome sequence is necessary to address comparative genomics questions such as structural variations among genotypes and gain or loss of specific function. Despite the spectacular progress that has been made in sequencing technologies, obtaining accurate and reliable data is still a challenge, both at the whole genome scale and when targeting specific genomic regions. These problems are even more noticeable for complex plant genomes. Most plant genomes are known to be particularly challenging due to their size, high density of repetitive elements and various levels of ploidy. To overcome these problems, we have developed a strategy to reduce genome complexity by using the large insert BAC libraries combined with next generation sequencing technologies.
Results:
We compared two different technologies (Roche-454 and Pacific Biosciences PacBio RS II) to sequence pools of BAC clones in order to obtain the best quality sequence. We targeted nine BAC clones from different species (maize, wheat, strawberry, barley, sugarcane and sunflower) known to be complex in terms of sequence assembly. We sequenced the pools of the nine BAC clones with both technologies. We compared assembly results and highlighted differences due to the sequencing technologies used.
Conclusions:
We demonstrated that the long reads obtained with the PacBio RS II technology serve to obtain a better and more reliable assembly, notably by preventing errors due to duplicated or repetitive sequences in the same region.
Link :
FRIZZY PANICLE drives supernumerary spikelets in bread wheat (T. aestivum L.).
Something went wrong when connecting to PubMed API.
Click here to access to the publication on PubMed website
FRIZZY PANICLE drives supernumerary spikelets in bread wheat (T. aestivum L.).
Plant Physiol. 2015 Jan;167(1):189-99. doi: 10.1104/pp.114.250043
Read more
Authors :
Dobrovolskaya O, Pont C, Sibout R, Martinek P, Badaeva E, Murat F, Chosson A, Watanabe N, Prat E, Gautier N, Gautier V, Poncet C, Orlov Y, Krasnikov A, Bergès H, Salina E, Laikova L, Salse J.
Plant Physiol. 2015 Jan;167(1):189-99. doi: 10.1104/pp.114.250043
Abstract :
Bread wheat (Triticum aestivum) inflorescences, or spikes, are characteristically unbranched and normally bear one spikelet per rachis node. Wheat mutants on which supernumerary spikelets (SSs) develop are particularly useful resources for work towards understanding the genetic mechanisms underlying wheat inflorescence architecture and, ultimately, yield components. Here, we report the characterization of genetically unrelated mutants leading to the identification of the wheat FRIZZY PANICLE (FZP) gene, encoding a member of the APETALA2/Ethylene Response Factor transcription factor family, which drives the SS trait in bread wheat. Structural and functional characterization of the three wheat FZP homoeologous genes (WFZP) revealed that coding mutations of WFZP-D cause the SS phenotype, with the most severe effect when WFZP-D lesions are combined with a frameshift mutation in WFZP-A. We provide WFZP-based resources that may be useful for genetic manipulations with the aim of improving bread wheat yield by increasing grain number.
Dominance hierarchy arising from the evolution of a complex small RNA regulatory network.
Something went wrong when connecting to PubMed API.
Click here to access to the publication on PubMed website
Dominance hierarchy arising from the evolution of a complex small RNA regulatory network.
Science. 346(6214):1200-5. doi: 10.1126/science.1259442.
Read more
Authors
Durand E1, Méheust R1, Soucaze M1, Goubet PM1, Gallina S1, Poux C1, Fobis-Loisy I2, Guillon E2, Gaude T2, Sarazin A3, Figeac M4, Prat E5, Marande W5, Bergès H5, Vekemans X1, Billiard S1, Castric V6.
Science. 346(6214):1200-5. doi: 10.1126/science.1259442.
Abstract
The prevention of fertilization through self-pollination (or pollination by a close relative) in the Brassicaceae plant family is determined by the genotype of the plant at the self-incompatibility locus (S locus). The many alleles at this locus exhibit a dominance hierarchy that determines which of the two allelic specificities of a heterozygous genotype is expressed at the phenotypic level. Here, we uncover the evolution of how at least 17 small RNA (sRNA)-producing loci and their multiple target sites collectively control the dominance hierarchy among alleles within the gene controlling the pollen S-locus phenotype in a self-incompatible Arabidopsis species. Selection has created a dynamic repertoire of sRNA-target interactions by jointly acting on sRNA genes and their target sites, which has resulted in a complex system of regulation among alleles.
Contrasted Patterns of Molecular Evolution in Dominant and Recessive Self-Incompatibility Haplotypes in Arabidopsis.
Something went wrong when connecting to PubMed API.
Click here to access to the publication on PubMed website
Contrasted Patterns of Molecular Evolution in Dominant and Recessive Self-Incompatibility Haplotypes in Arabidopsis.
PLoS Genet.2012 Mar;8(3):e1002495.
PMID: 22457631
Read more
Authors
Goubet PM, Bergès H, Bellec A, Prat E, Helmstetter N, Mangenot S, Gallina S, Holl AC, Fobis-Loisy I, Vekemans X, Castric V.
PLoS Genet. 2012 Mar;8(3):e1002495.
PMID: 22457631
Abstract
Self-incompatibility has been considered by geneticists a model system for reproductive biology and balancing selection, but our understanding of the genetic basis and evolution of this molecular lock-and-key system has remained limited by the extreme level of sequence divergence among haplotypes, resulting in a lack of appropriate genomic sequences. In this study, we report and analyze the full sequence of eleven distinct haplotypes of the self-incompatibility locus (S-locus) in two closely related Arabidopsis species, obtained from individual BAC libraries. We use this extensive dataset to highlight sharply contrasted patterns of molecular evolution of each of the two genes controlling self-incompatibility themselves, as well as of the genomic region surrounding them. We find strong collinearity of the flanking regions among haplotypes on each side of the S-locus together with high levels of sequence similarity. In contrast, the S-locus region itself shows spectacularly deep gene genealogies, high variability in size and gene organization, as well as complete absence of sequence similarity in intergenic sequences and striking accumulation of transposable elements. Of particular interest, we demonstrate that dominant and recessive S-haplotypes experience sharply contrasted patterns of molecular evolution. Indeed, dominant haplotypes exhibit larger size and a much higher density of transposable elements, being matched only by that in the centromere. Overall, these properties highlight that the S-locus presents many striking similarities with other regions involved in the determination of mating-types, such as sex chromosomes in animals or in plants, or the mating-type locus in fungi and green algae.
Advancing Eucalyptus genomics: identification and sequencing of lignin biosynthesis genes from deep-coverage BAC libraries
Something went wrong when connecting to PubMed API.
Click here to access to the publication on PubMed website
Advancing Eucalyptus genomics: identification and sequencing of lignin biosynthesis genes from deep-coverage BAC libraries
BMC Genomics. 2011 Mar 4;12(1):137.
Read more
Authors :
Paiva JA, Prat E, Vautrin S, Santos MD, San-Clemente H, Brommonschenkel S, Fonseca PG, Grattapaglia D, Song X, Ammiraju JS, Kudrna D, Wing RA, Freitas AT, Berges H, Grima-Pettenati J.
BMC Genomics. 2011 Mar 4;12(1):137.
Abstract :
BACKGROUND: Eucalyptus species are among the most planted hardwoods in the world because of their rapid growth, adaptability and valuable wood properties. The development and integration of genomic resources into breeding practice will be increasingly important in the decades to come. Bacterial artificial chromosome (BAC) libraries are key genomic tools that enable positional cloning of important traits, synteny evaluation, and the development of genome framework physical maps for genetic linkage and genome sequencing.
RESULTS: We describe the construction and characterization of two deep-coverage BAC libraries EG_Ba and EG_Bb obtained from nuclear DNA fragments of E. grandis (clone BRASUZ1) digested with HindIII and BstYI, respectively. Genome coverages of 17 and 15 haploid genome equivalents were estimated for EG_Ba and EG_Bb, respectively. Both libraries contained large inserts, with average sizes ranging from 135Kb (Eg_Bb) to 157Kb (Eg_Ba), very low extra-nuclear genome contamination providing a probability of finding a single copy gene [greater than or equal to] 99.99%. Libraries were screened for the presence of several genes of interest via hybridizations to high-density BAC filters followed by PCR validation. Five selected BAC clones were sequenced and assembled using the Roche GS FLX technology providing the whole sequence of the E. grandis chloroplast genome, and complete genomic sequences of important lignin biosynthesis genes.
CONCLUSIONS: The two E. grandis BAC libraries described in this study represent an important milestone for the advancement of Eucalyptus genomics and forest tree research. These BAC resources have a highly redundant genome coverage (>15x), contain large average inserts and have a very low percentage of clones with organellar DNA or empty vectors. These publicly available BAC libraries are thus suitable for a broad range of applications in genetic and genomic research in Eucalyptus and possibly in related species of Myrtaceae, including genome sequencing, gene isolation, functional and comparative genomics. Because they have been constructed using the same tree (E. grandis BRASUZ1) whose full genome is being sequenced, they should prove instrumental for assembly and gap filling of the upcoming Eucalyptus reference genome sequence.
Construction and characterization of two BAC libraries representing a deep-coverage of the genome of chicory (Cichorium intybus L., Asteraceae).
Something went wrong when connecting to PubMed API.
Click here to access to the publication on PubMed website
Construction and characterization of two BAC libraries representing a deep-coverage of the genome of chicory (Cichorium intybus L., Asteraceae).
BMC Res Notes. 2010 Aug 11;3:225.
Read more
Abstract
BACKGROUND: The Asteraceae represents an important plant family with respect to the numbers of species present in the wild and used by man. Nonetheless, genomic resources for Asteraceae species are relatively underdeveloped, hampering within species genetic studies as well as comparative genomics studies at the family level. So far, six BAC libraries have been described for the main crops of the family, i.e. lettuce and sunflower. Here we present the characterization of BAC libraries of chicory (Cichorium intybus L.) constructed from two genotypes differing in traits related to sexual and vegetative reproduction. Resolving the molecular mechanisms underlying traits controlling the reproductive system of chicory is a key determinant for hybrid development, and more generally will provide new insights into these traits, which are poorly investigated so far at the molecular level in Asteraceae.
FINDINGS: Two bacterial artificial chromosome (BAC) libraries, CinS2S2 and CinS1S4, were constructed from HindIII-digested high molecular weight DNA of the contrasting genotypes C15 and C30.01, respectively. C15 was hermaphrodite, non-embryogenic, and S2S2 for the S-locus implicated in self-incompatibility, whereas C30.01 was male sterile, embryogenic, and S1S4. The CinS2S2 and CinS1S4 libraries contain 89,088 and 81,408 clones. Mean insert sizes of the CinS2S2 and CinS1S4 clones are 90 and 120 kb, respectively, and provide together a coverage of 12.3 haploid genome equivalents. Contamination with mitochondrial and chloroplast DNA sequences was evaluated with four mitochondrial and four chloroplast specific probes, and was estimated to be 0.024% and 1.00% for the CinS2S2 library, and 0.028% and 2.35% for the CinS1S4 library. Using two single copy genes putatively implicated in somatic embryogenesis, screening of both libraries resulted in detection of 12 and 13 positive clones for each gene, in accordance with expected numbers.
CONCLUSIONS: This indicated that both BAC libraries are valuable tools for molecular studies in chicory, one goal being the positional cloning of the S-locus in this Asteraceae species.
Authors
Gonthier L, Bellec A, Blassiau C, Prat E, Helmstetter N, Rambaud C, Huss B, Hendriks T, Bergès H, Quillet MC.
BMC Res Notes. 2010 Aug 11;3:225.
Something went wrong when connecting to PubMed API.
Click here to access to the publication on PubMed website


