Ger4 locus characterization in 8 barley varieties and it relation with powdery mildew resistance

Project coordinator:
Patrick Schweizer, Dr.
Head of Genome Analysis program
Head of Pathogen-Stress Genomics lab
Leibniz-Institut für Pflanzengenetik und Kulturpflanzenforschung (IPK)
Genomzentrum
Corrensstrasse 3
D-06466 Gatersleben
Germany
Abstract:
Immunity of plants triggered by pathogen-associated molecular patterns (PAMPs) is based on the execution of an evolutionarily conserved defense response that includes the accumulation of pathogenesis-related (PR) proteins as well as multiple other defenses. The most abundant PR transcript of barley (Hordeum vulgare) leaf epidermis attacked by the powdery mildew fungus Blumeria graminis f. sp hordei encodes the germin-like protein GER4, which has superoxide dismutase activity and functions in PAMP-triggered immunity.
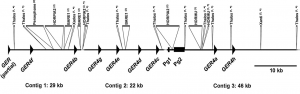
Patrick Schweizer ‘s team (IPK- Germany) showed that barley GER4 is encoded by a dense cluster of tandemly duplicated genes (GER4a-h) that underwent several cycles of duplication. The genomic organization of the GER4 locus also provides evidence for repeated gene birth and death cycles. They also identify 8 barley varieties that show different GER4 expression and different Blumeria graminis f. sp hordei resistance pattern.
The goal of the project is to obtain the GER4 locus sequence of those varieties.
Barley genome is around 5.1 Gb and is mainly composed by repeated sequences (>80 %). In front of this complexity, building and screening non-gridded BAC library was the best approach to obtain this region with a high sequence quality that is necessary to make sequence comparison.
At the CNRGV, we are building and screening the 8 BAC libraries in order to identify the GER4 locus that is around 100kb long in size.
|
|
|
CNRGV's responsible
William Marande