The CNRGV has recently acquired a P2 solo from Oxford Nanopore Technologies in the frame of the GREASE project funded by Plant2Pro. This benchtop sequencer offers an easy library preparation and loading, with an expected throughput of 150Gb per flow-cell.
To assess the genome assembly quality we can expect from ONT data, we sequenced one bean genotype (Phaseolus vulgaris, estimated genome size 600Mb) on a multiplexed flow-cell. Of the 150Gb of data produced, 52Gb were generated for the bean genome with a read N50 of 18kb and a maximum read length of 387kb. We assembled the data using Flye tool and finally obtained 224 contigs for a total length of 538 Mb, a N50 of 12Mb and a very good BUSCO score of 99.2%.
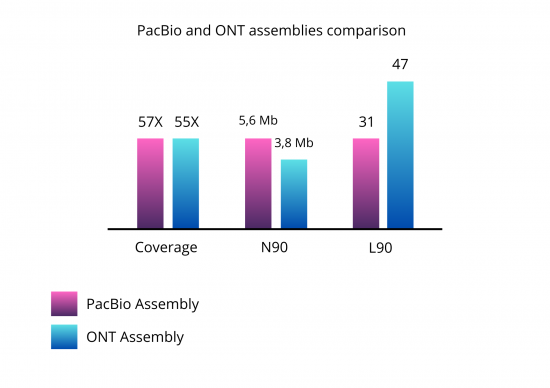
This bean genotype has previously been sequenced using PacBio, known as very high fidelity long reads technology. The results of the two assemblies are compared in the graph below:
With a similar coverage of 55X, we obtain 2 assemblies of 560 and 540Mb. The ONT assembly is more fragmented (224 vs 83 contigs) but 90% of the genome is represented in the 47 largest contigs and the Busco score is equivalent to the PacBio assembly.
To conclude, based on these tests, ONT genome assemblies are similar to PacBio in terms of accuracy and completeness. Although they remain more fragmented, the assemblies are excellent quality. ONT appears as an efficient and reliable solution to sequence small and homozygous plant genomes. For larger, heterozygous genomes, PacBio remains the reference technology. To achieve telomere to telomere assembly, a complementary scaffolding method such as Hi-C or optical map remains essential to obtain the best results.