Gene@Home - Newsletter June 2010
CNRGV, The French Plant Genomic Resource Center is both a service provider and repository of plant genomes and data. Based in Toulouse, It's a unique service center for plant genomics in France.
New Genomic Libraries available:
|
|
|
|
New services:
BAC library construction
We produce genomic libraries on demand, from DNA extraction to colony picking. We propose different strategies according to the level of genomic data available. Various libraries have been constructed in the frame of collaboration with several laboratories.
| Common name | Library |
| Arabidopsis Halleri |
Aha-B-HF11 |
| Arabidopsis Lyrata |
Aly-B-03A7 |
| Arabidopsis thaliana | Ath-B-ITA |
| Chicory |
Cin-B-S1 |
| Sunflower |
Han-B-XRQ |
| Medicago | Mtr-B-F83 |
| Passiflora | Ped-B-Flav |
| Spartine | Sma-B-MOB |
In collaboration with various laboratories, we are also setting up a new approach in order to isolate genomic regions of interest in specific genotype background. The main aim of this approach is to rapidly have access to genomic regions involved in specific biological process by using non gridded BAC libraries.
Screening services
New DNA pools are now available for screening and distribution :
| Library | Pool Producer | Type of pools | Total Coverage | Number of PCR reactions per screening |
| Sunflower Han-B-XRQ | CNRGV | 3D | 4X | 16 + 74 per positive block (~0,3X) |
| Sugarcane Shy-B-R570 | CNRGV | 3D | 1.3X | 14 + 66 per positive block (~0,4X) |
| Durum Wheat Ttu-B-LDN65 | CIRAD MONTPELLIER / CNRGV | 2D | 5X | 292 + 42 per positive plate |
| Bread Wheat TaaCsp1AShA | CNRGV | 3D | 1X (MTP) * | 50 |
| Bread Wheat TaaCsp1BShA | CNRGV | 3D | 1X (MTP) * | 60 |
| Bread Wheat TaaCsp3BFh_MTPv2 | CNRGV | 3D | 1X (MTP) * | 62 |
* MTP = Pooling of the clones of the Wheat Chromosome Minimal Tiling Path
Macroarrays production > New pattern
BAC clones of a library can be spotted using a 7*7 pattern. This density allows a significant decrease of the number of filters to be hybridized in order to perform a screening. You can spot 110 592 duplicated BAC clones (144 microplates in duplicate) on a single membrane.
Macroarrays already available using a 7*7 pattern:
| Common name | Library |
| Arabidopsis lyrata | Aly-B-05B8 |
| Chicory | Cin-B-S1 |
| Chicory | Cin-B-S2 |
| Barley | HVVMRXALLhA |
| Barley | HVVMRXALLhB |
| Barley | HVVMRXALLrA |
| Barley | HVVMRXALLmA |
| Barley | HVVMRXALLeA |
| Rye | Sce-B-FRG Blanco |
| Surgarcane | Shy-B-R570 |
| Bread Wheat | Tae-B-Renan |
| Durum Wheat | Ttu-B-LDN65 |
| Maize | Biogemma F2 BAC |
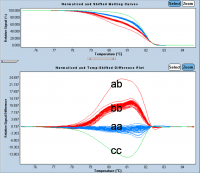
HRM genotyping
|
We now offer a service of HRM genotyping. This high throughput genotyping method is based on a classical PCR run on Real Time PCR LC480 ROCHE (96 or 384 well format). The PCR is followed by a High Resolution Melting Analysis of the amplified fragments, allowing discrimination of different alleles, up to one SNP variability between two genotypes. |
|
New collaborations :
|
|
The CNRGV is the partner of the genome sequencing project involving Canada, USA and France. In this project, the CNRGV will be in charge of the constructions of the new sunflower BAC libraries from the genotype HA412-HO. |
|
The Sugarcane Genome Sequencing Initiative (SUGESI): Strategies for Sequencing a Highly Complex Genome. |
|
|
Construction and screening a barley BAC library in order to isolate the genomic region responsible for the rust resistance phenotype. |
|
Thesis on "Genetic-chromosomic maps of Passiflora". The objective of this study is to construct a new integrated molecular map combining different loci configuration. |
Publications
Brassica orthologs from BANYULS belong to a small multigene family, which is involved in procyanidin accumulation in the seed
Read more...
Fine Mapping and Marker Development for the Crossability Gene SKr on Chromosome 5BS of Hexaploid Wheat (Triticum aestivum L.)
Read more...
ISO 9001:2008 certification
| The CNRGV has renewed its ISO 9001:2008 certification by the SGS certifier organization. Its certification covers all its activities: Centralisation, validation and distribution of plant genomic resources, genomic tools and services for studying plant genomic collections. |
|