Non gridded BAC library
|
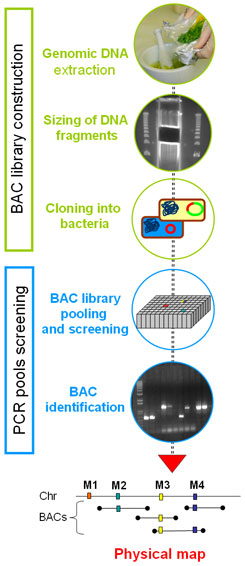
A BAC library provides long-term efficacy in genome analysis. Its versatility of use, such as map-based cloning, physical mapping or sequencing, facilitates multi-faceted projects. However, the construction of a BAC library is great and undertaking if it only aims to know one or even a few genomic regions.
In order to reduce costs and improve access to genomic regions of interest, the CNRGV has developed a targeted genomic library strategy. BAC clones of targeted genomic libraries are organized in pools containing around 1000 to 3000 events and screened immediately after their transformation. Thus allows for the isolation of clones of interest, while avoiding the expensive and time-consuming step of organizing BAC clones in microplates.
The CNRGV offers the possibility to construct a targeted BAC library from a genotype of interest, starting with a 1 or 2 x genome coverage (with mean inserts size around 110kb). The pools are then screened using specific markers of the region of interest, using real-time PCR. The BAC clones carrying the markers are then individualized and can be sequenced using Next Generation DNA Sequencing technology (pool of BACs with Pacific Biosciences technology).
Targeted genomic libraries have proven efficacy and are notably useful in investigating the diversity of specific regions among various plant cultivars. |
|
Various projects are finished or in progress through collaboration:
- Ger4 locus characterization in 8 barley varieties and it relation with powdery mildew resistance (IPK, Germany)
- ZeaWall : Towards the identification of genetic determinisms involved in lignified secondary cell wall degradability through the elucidation of QTLs (Crop companies)
- Mapping of the tiller inhibition gene tin for Oligoculm wheat line / Jérôme SALSE (INRA-Clermont Ferrand, France) and Wolfgang SPIELMEYER, (CSIRO-Canberra, Australia)
- Characterization of a genomic region of interest in Courtot wheat line / Pierre SOURDILLE - Catherine FEUILLET (INRA-Clermont Ferrand)
- Characterization of sr2 locus in wheat Hope line involved in durable resistance to the wheat stem rust pathogen / Wolfgang SPIELMEYER (CSIRO-Canberra, Australia) / http://www.ncbi.nlm.nih.gov/pubmed/25547135
- Characterization of a genomic region of interest in Pavon 76 wheat line / Evans LAGUDAH (CSIRO-Canberra, Australia)
- Identification of the genomic region responsible for the resistance to the leaf rust in the weat variety Sinvalocho MA using non gridded BAC library.
- Characterization of a genomic region of interest in barley / Jan GIELEN - Carine Rizzolatti (Syngenta-Toulouse, France)
The targeted BAC library strategy is an efficient way to isolate your genomic region of interest and to explore the variability among specific genotypes / plant cultivar.

 Genomic samples storage & distribution
Genomic samples storage & distribution Large DNA Fragment cloning and characterization
Large DNA Fragment cloning and characterization HMW DNA extraction
HMW DNA extraction Genome Scaffolding Hi-C / Bionano
Genome Scaffolding Hi-C / Bionano Genome sequencing and Assembly
Genome sequencing and Assembly Transcriptome & Annotation
Transcriptome & Annotation Target sequencing
Target sequencing